Cells make Z-RNAs as the first line of defense against pathogenic viruses
Viruses attack by taking control of a cell. To avoid becoming a virus factory, infected cells trigger rapid cell death by producing Z-RNAs made from "junk" DNA.
Once controversial, it is now widely accepted that Z-DNA and Z-RNA play essential roles in normal cellular biology. When deregulated, these pathways exacerbate many chronic inflammatory diseases”
CHARLESTOWN, MA, UNITED STATES, October 13, 2025 /EINPresswire.com/ -- In a paper appearing in the journal Nature today, a multinational team of scientists confirms the ancient role of Z-RNA in defending the host against viruses. This unusual left-handed RNA structure is sensed by the ZBP1 protein that recognizes Z-RNA through the conformation-specific Zα domain. ZBP1 then triggers inflammatory cell death. The destruction of the cell prevents its conversion into a viral factory. The response alerts the adaptive immune system, which then proceeds to kill other virally infected cells.— Alan Herbert
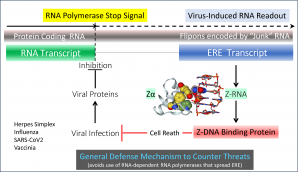
The source of the Z-RNAs is “junk” DNA. Previously, “junk” DNA has been considered non-functional as it lacks informational values, given its limited sequence diversity and high frequency throughout the genome. This “junk” has accumulated through the spread of endogenous retroelements (ERE) that copy and paste themselves from one chromosomal locus to another. EREs contain repetitive sequences, called flipons, that can form different DNA and RNA structures. This transformation occurs without any change to the DNA sequence or break in the DNA backbone. This change in conformation leads to the assembly of structure-specific protein complexes that initiate different outcomes. The cell can then respond rapidly to changing conditions by sensing the switch in flipon structure.
During viral infection, Z-RNAs are recognized by Z-DNA binding Protein 1, which has two structure-specific Zα domains. The only other protein in the human genome with a Zα, the double-stranded editing enzyme ADAR, negatively regulates immune responses against normal host-encoded RNAs. In contrast to ADAR, ZBP1 is pro-inflammatory. The protein induces cell death when ADAR is unable to suppress its activation by Z-RNAs. This system of regulation dates back to the first single-cell progenitor of modern multicellular organisms that enclose their DNA within a nucleus.
How then does a cell know how to produce Z-RNAs during a viral infection? The mechanism involved is detailed in the Nature paper (Figure). It exploits the manner in which Herpes and Influenza viruses take over the cell. One of the first steps is to shut down the readout of host genes so that the cell cannot make proteins to thwart the viral attack. Both viruses achieve this by trapping the host RNA polymerase enzyme on the host DNA, thereby disrupting the normal process of transcript termination. Instead of halting RNA polymerization and being released from DNA, the RNA-making enzymes continue onwards, producing extremely long transcripts. The polymerase then acts as a plow, displacing the protein complexes needed to read out downstream genes and to process their transcripts properly. The process of shutting down host transcription is usually complete 2-6 hours after the first virus enters the cell. Subsequently, the virus utilizes the cell ghost to produce many new copies of itself. The viral factory furthers the infection and the propagation of progeny to new hosts.
The cell, therefore, has only a short time to halt the virus infection before it takes control. In a classic case of an evolutionary arms race, the cell sets a trap that triggers the production of Z-RNAs. By retaining Z-RNA-forming flipons encoded by EREs within “junk’ DNA that lies just beyond the normal termination sites, the cell can immediately make the Z-RNAs needed to trigger cell death as soon as the virus interferes with the release of the RNA polymerase from DNA. The paper reveals that the multitude of Z-RNAs produced by the disruption of transcript termination (DoTT) is sufficient to cause the death of host cells.
The work further demonstrates that expression of the viral proteins that induce DoTT does indeed induce Z-RNA production and is sufficient to trigger cell death. Furthermore, the clinically used drug JTE-607, which is highly active against blood cancers, including leukemia and Ewing’s sarcoma, also induces Z-RNAs by interfering with transcript termination. The authors also note that any mechanism that dysregulates transcription in a cell is also likely to result in production of Z-RNA from ERE flipons and induction of inflammatory responses. Such outcomes may arise during cell stress of any type and also in senescent cells.
What are the origins of this defense mechanism? The dispersion of ERE throughout the genome depends on an enzyme called reverse transcriptase (RT) that copies ERE transcripts back into DNA. Round worms, a favorite genetic model organism, largely eliminated EREs. Then they were able to amplify pathogen-specific RNAs using host-encoded RTs. The risk of promoting the spread of ERE by RTs throughout their genome was sufficiently low compared with the benefit of spreading the small RNAs to cells and to embryos to provide host protection against pathogens. Other repurposed EREs and the flipons they encoded as these sequences endowed them with much greater phenotypic diversity – different outcomes could be generated by changing the conformation of these sequences. Indeed, many regulatory sequences in human development and differentiation are ERE derived and exploit the ability of flipons to switch programs on and off. The trade-off was to avoid using host-encoded RTs that might further ERE spread haphazardly throughout the genome. Instead, the ERE encoded Z-RNAs were used to trigger host defenses against pathogens.
The corresponding authors are Sid Balachandran from Fox Case Cancer Center, Alan Herbert from InsideOutBio, Ting Zhang from Sichuan University, and Lars Dölken from the Technical University of Munich. Bioinformatic analysis was performed by a group headed by Maria Poptsova, HSE in Moscow, and sequencing at St Jude's Hospital under the guidance of Paul Thomas. Dr. Herbert, a founder of InsideOutBio, discovered the Zα domain and has since provided genetic evidence for the role of flipons in regulating various cellular transactions.
InsideOutBio is a start-up focused on developing a novel class of proprietary therapeutics to ‘light up tumors for the immune system. These statements about InsideOutBio comply with Safe-Harbor laws. They are forward-looking and involve known and unknown risks and uncertainties. They are not guarantees of future performance, and undue reliance should not be placed on them.
Alan Herbert
InsideOutBio, Inc
+1 617-584-0360
email us here
Legal Disclaimer:
EIN Presswire provides this news content "as is" without warranty of any kind. We do not accept any responsibility or liability for the accuracy, content, images, videos, licenses, completeness, legality, or reliability of the information contained in this article. If you have any complaints or copyright issues related to this article, kindly contact the author above.